Can you carry out a quantified risk of reidentification on documents when you don't have access to the underlying datasets?
I get asked this question every year, either during a presentation or whilst its my turn to man the booth at a conference. It always really pained me to say “No, I'm afraid Blur can only be used for redactions when there is no underlying data”. Within our CTT services business we have never had a need to explore any other option, as all our sponsors did have access to the data…… until just recently.
We were approached by a sponsor to anonymize some documents for a regulatory submission. We explained to them our process of risk measurement and anonymization, and they asked the same question above. Rather than say “No” though, we actually had a “Maybe”. Using a Python script we were able to mine the documents for several identifiers. The documents came to us, complete with 16.2 listings (which are of course out of scope for a Health Canada PRCI submission).
But this meant that we had the data, just in PDF form rather than the standard SAS dataset format we're more familiar with. We just had to extract the data from the PDFs into something more usable.
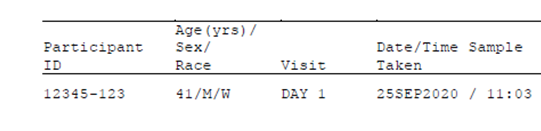
Above is a screen shot of the format from one of the listings (Populated with fake data of course).
If you export this into a Text (Accessible) format its looks a bit like this:
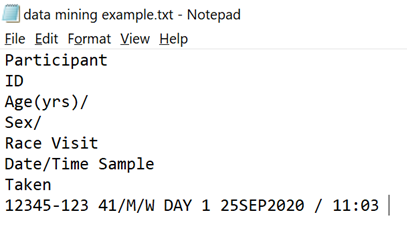
The title information looks very messy, but the actual patient data is very easy to read, and a very specific format. Patient ID of 5 digits a hyphen and 3 digits followed by a space then age forward slash gender forward slash race.
So now we can use a regular expression to mine this information from the document.
“\d\d\d\d\d-\d\d\d \d\d/\w/\w”
This returns a list of all the subject ids followed by age, sex and race that appeared in this format throughout all documents in the submission.
We then searched the documents for just the subject IDs alone.
“\d\d\d\d\d-\d\d\d”
This returned many more occurrences of the subject IDs, as of course they appeared in narratives as well as the listings. However, all the subject IDs were checked against our first set, so we could make sure we had a complete list of all patients IDs that appear in the document as well as the relevant identifying information.
Everyone on the trial was from the same country and there was no mention of ethnicity, height, weight or BMI anywhere in the documents.
After extracting this data with a small Python script, I used a small SAS program to create the underlying risk dataset that Blur needs to anonymize documents.
→ Request the scripts used
This meant that we could run some simulations using k-anonymity, and rather than redacting age sex and race we could keep sex, apply a low frequency redact to race, and apply a numeric band to age. We could also offset all the patient dates now that we had a list of patient IDs. This created anonymized documents with much higher utility than simply redacted documents.
Using this method, we could also supply a risk of reidentification score.
In conclusion, when someone now asks me this question, I do still answer “Blur needs underlying data to carry out a risk of reidentification score. However, the required data may be able to be mined from the document with some extra effort.”
Document & Data Sharing for Life Sciences
Take advantage of an on-demand, flexible outsource service designed to measure risk and anonymize data and documents within strict sharing deadlines. Leveraging Blur anonymization software, our team of transformation experts provide swift data and document anonymization and quantifiable risk measurements so your organization can confidently share on any scale.
Contact us to get your team started today.